Introduction
This exercise focuses on conducting exploratory data analysis from the influenza data.
The raw data for this exercise comes from the following citation: McKay, Brian et al. (2020), Virulence-mediated infectiousness and activity trade-offs and their impact on transmission potential of patients infected with influenza, Dryad, Dataset, https://doi.org/10.5061/dryad.51c59zw4v.
The processed data was produced on the Data Processing tab.
Within this analysis, the main continuous outcome of interest is body temperature and the main categorical outcome of interest is nausea. For each symptom, we will do the following steps:
- Produce and print some numerical output (e.g. table, summary statistics)
- Create a histogram or density plot (continuous variables only)
- Scatterplot, boxplot, or other similar plots against the main outcome of interest
- Any other exploration steps that may be useful.
For each variable, the EDA steps will be labeled by the numbers listed above.
Required Packages
The following R packages are required for this exercise:
- here: for path setting
- tidyverse: for all packages in the Tidyverse (ggplot2, dyplr, tidyr, readr, purr, tibble, stringr, forcats)
- summarytools: for overall dataframe summary
- ggplot2: for plotting data
- car: for creating QQ plots
- table1: for creating tables for summary statistics / other numerical outputs
- scales: for percent calculation
- margrittr: for sequential piping
Load Processed Data
Load the data created on the Data Processing page.
# path to data
# note the use of the here() package and not absolute paths
data_location <- here::here("data","flu","processeddata.rds")
# load data using the "ReadRDS" function in base R.
processeddata <- base::readRDS(data_location)
# create a mirror dataset to avoid manipulating the cleaned dataset
EDAdata <- processeddata
Data Overview
To better understand the processed data, let’s use summarytools to visualize the dataframe.
summarytools::dfSummary(EDAdata)
## Data Frame Summary
## Dimensions: 730 x 32
## Duplicates: 0
##
## -----------------------------------------------------------------------------------------------------------------
## No Variable Stats / Values Freqs (% of Valid) Graph Valid Missing
## ---- ------------------- ------------------------ -------------------- --------------------- ---------- ---------
## 1 SwollenLymphNodes 1. No 418 (57.3%) IIIIIIIIIII 730 0
## [factor] 2. Yes 312 (42.7%) IIIIIIII (100.0%) (0.0%)
##
## 2 ChestCongestion 1. No 323 (44.2%) IIIIIIII 730 0
## [factor] 2. Yes 407 (55.8%) IIIIIIIIIII (100.0%) (0.0%)
##
## 3 ChillsSweats 1. No 130 (17.8%) III 730 0
## [factor] 2. Yes 600 (82.2%) IIIIIIIIIIIIIIII (100.0%) (0.0%)
##
## 4 NasalCongestion 1. No 167 (22.9%) IIII 730 0
## [factor] 2. Yes 563 (77.1%) IIIIIIIIIIIIIII (100.0%) (0.0%)
##
## 5 CoughYN 1. No 75 (10.3%) II 730 0
## [factor] 2. Yes 655 (89.7%) IIIIIIIIIIIIIIIII (100.0%) (0.0%)
##
## 6 Sneeze 1. No 339 (46.4%) IIIIIIIII 730 0
## [factor] 2. Yes 391 (53.6%) IIIIIIIIII (100.0%) (0.0%)
##
## 7 Fatigue 1. No 64 ( 8.8%) I 730 0
## [factor] 2. Yes 666 (91.2%) IIIIIIIIIIIIIIIIII (100.0%) (0.0%)
##
## 8 SubjectiveFever 1. No 230 (31.5%) IIIIII 730 0
## [factor] 2. Yes 500 (68.5%) IIIIIIIIIIIII (100.0%) (0.0%)
##
## 9 Headache 1. No 115 (15.8%) III 730 0
## [factor] 2. Yes 615 (84.2%) IIIIIIIIIIIIIIII (100.0%) (0.0%)
##
## 10 Weakness 1. None 49 ( 6.7%) I 730 0
## [factor] 2. Mild 223 (30.5%) IIIIII (100.0%) (0.0%)
## 3. Moderate 338 (46.3%) IIIIIIIII
## 4. Severe 120 (16.4%) III
##
## 11 WeaknessYN 1. No 49 ( 6.7%) I 730 0
## [factor] 2. Yes 681 (93.3%) IIIIIIIIIIIIIIIIII (100.0%) (0.0%)
##
## 12 CoughIntensity 1. None 47 ( 6.4%) I 730 0
## [factor] 2. Mild 154 (21.1%) IIII (100.0%) (0.0%)
## 3. Moderate 357 (48.9%) IIIIIIIII
## 4. Severe 172 (23.6%) IIII
##
## 13 CoughYN2 1. No 47 ( 6.4%) I 730 0
## [factor] 2. Yes 683 (93.6%) IIIIIIIIIIIIIIIIII (100.0%) (0.0%)
##
## 14 Myalgia 1. None 79 (10.8%) II 730 0
## [factor] 2. Mild 213 (29.2%) IIIII (100.0%) (0.0%)
## 3. Moderate 325 (44.5%) IIIIIIII
## 4. Severe 113 (15.5%) III
##
## 15 MyalgiaYN 1. No 79 (10.8%) II 730 0
## [factor] 2. Yes 651 (89.2%) IIIIIIIIIIIIIIIII (100.0%) (0.0%)
##
## 16 RunnyNose 1. No 211 (28.9%) IIIII 730 0
## [factor] 2. Yes 519 (71.1%) IIIIIIIIIIIIII (100.0%) (0.0%)
##
## 17 AbPain 1. No 639 (87.5%) IIIIIIIIIIIIIIIII 730 0
## [factor] 2. Yes 91 (12.5%) II (100.0%) (0.0%)
##
## 18 ChestPain 1. No 497 (68.1%) IIIIIIIIIIIII 730 0
## [factor] 2. Yes 233 (31.9%) IIIIII (100.0%) (0.0%)
##
## 19 Diarrhea 1. No 631 (86.4%) IIIIIIIIIIIIIIIII 730 0
## [factor] 2. Yes 99 (13.6%) II (100.0%) (0.0%)
##
## 20 EyePn 1. No 617 (84.5%) IIIIIIIIIIIIIIII 730 0
## [factor] 2. Yes 113 (15.5%) III (100.0%) (0.0%)
##
## 21 Insomnia 1. No 315 (43.2%) IIIIIIII 730 0
## [factor] 2. Yes 415 (56.8%) IIIIIIIIIII (100.0%) (0.0%)
##
## 22 ItchyEye 1. No 551 (75.5%) IIIIIIIIIIIIIII 730 0
## [factor] 2. Yes 179 (24.5%) IIII (100.0%) (0.0%)
##
## 23 Nausea 1. No 475 (65.1%) IIIIIIIIIIIII 730 0
## [factor] 2. Yes 255 (34.9%) IIIIII (100.0%) (0.0%)
##
## 24 EarPn 1. No 568 (77.8%) IIIIIIIIIIIIIII 730 0
## [factor] 2. Yes 162 (22.2%) IIII (100.0%) (0.0%)
##
## 25 Hearing 1. No 700 (95.9%) IIIIIIIIIIIIIIIIIII 730 0
## [factor] 2. Yes 30 ( 4.1%) (100.0%) (0.0%)
##
## 26 Pharyngitis 1. No 119 (16.3%) III 730 0
## [factor] 2. Yes 611 (83.7%) IIIIIIIIIIIIIIII (100.0%) (0.0%)
##
## 27 Breathless 1. No 436 (59.7%) IIIIIIIIIII 730 0
## [factor] 2. Yes 294 (40.3%) IIIIIIII (100.0%) (0.0%)
##
## 28 ToothPn 1. No 565 (77.4%) IIIIIIIIIIIIIII 730 0
## [factor] 2. Yes 165 (22.6%) IIII (100.0%) (0.0%)
##
## 29 Vision 1. No 711 (97.4%) IIIIIIIIIIIIIIIIIII 730 0
## [factor] 2. Yes 19 ( 2.6%) (100.0%) (0.0%)
##
## 30 Vomit 1. No 652 (89.3%) IIIIIIIIIIIIIIIII 730 0
## [factor] 2. Yes 78 (10.7%) II (100.0%) (0.0%)
##
## 31 Wheeze 1. No 510 (69.9%) IIIIIIIIIIIII 730 0
## [factor] 2. Yes 220 (30.1%) IIIIII (100.0%) (0.0%)
##
## 32 BodyTemp Mean (sd) : 98.9 (1.2) 57 distinct values : 730 0
## [numeric] min < med < max: : : (100.0%) (0.0%)
## 97.2 < 98.5 < 103.1 : :
## IQR (CV) : 1.1 (0) : : :
## : : : : : . . . .
## -----------------------------------------------------------------------------------------------------------------
Looking at the data summary, there are multiple variables for the same symptom (e.g. category and presence). Additionally, several of the categorical variables have an uneven proportion distribution, and BodyTemp appears to have some skew as well.
The outcome variables of interest were specified as part of the MADA course, so what predictor varaibles may be relevant?
- The data is about influenza, so certainly symptoms commonly associated with influeza (e.g. runny nose, nasal congestion, chills / sweating, and myalgia).
- Since we are interested in nausea, we should also probably include nausea and vomiting (co-presentation).
Main Continuous Outcome of Interest: Body Temperature
# start with main continuous outcome of interest: body temperature
# (1) since it is continuous, we can calculate summary statistics with the base summary function
base::summary(EDAdata$BodyTemp)
## Min. 1st Qu. Median Mean 3rd Qu. Max.
## 97.20 98.20 98.50 98.94 99.30 103.10
# (2) create a density plot for body temperature
ggplot2::ggplot(data = EDAdata, aes(x = BodyTemp)) +
geom_density()
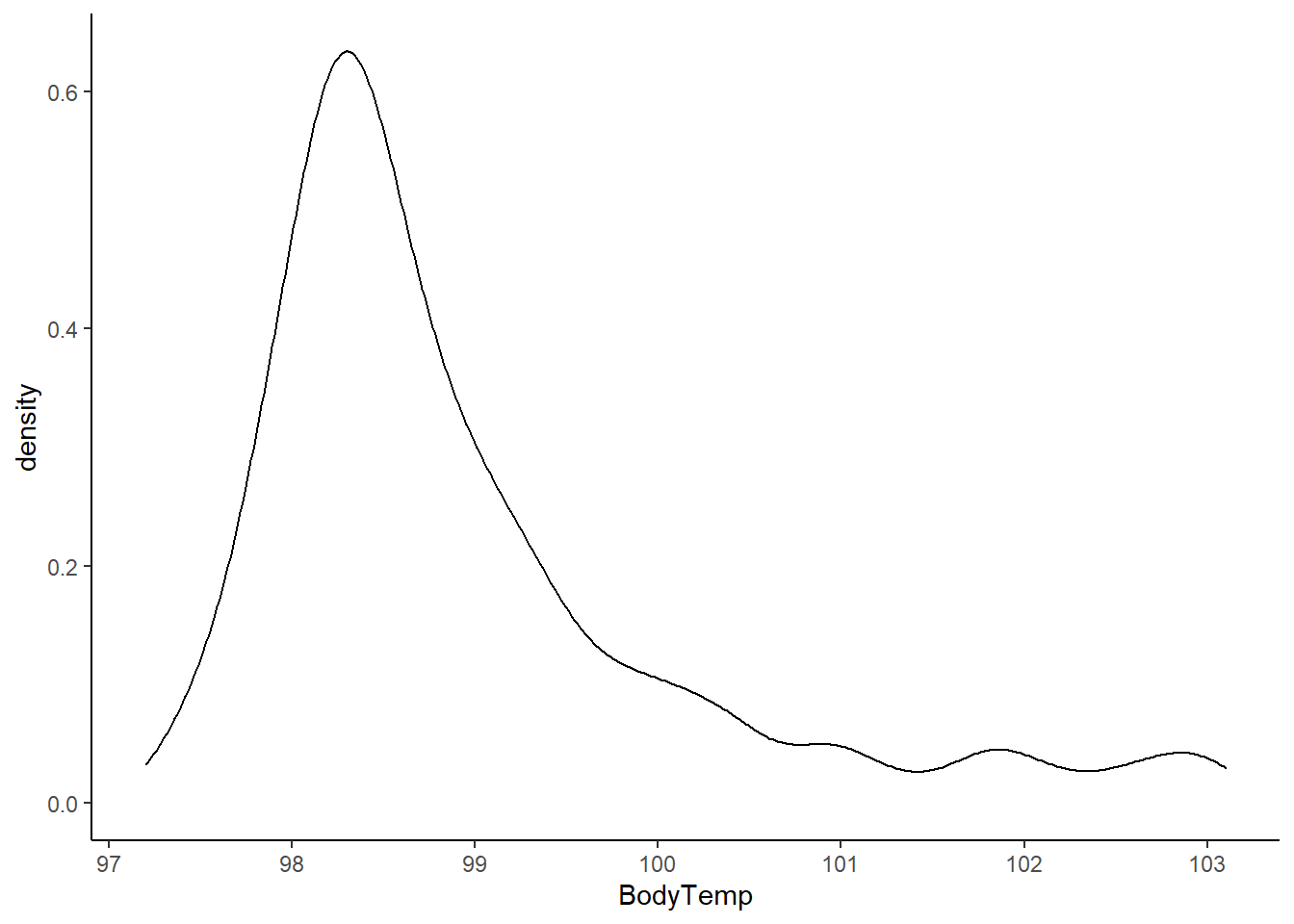
# looking at the histogram and summary statistics, there appears to be a left skew from a normal distribution
# (3) create a boxplot to better visualize
# since body temperature is the main outcome of interest, no need to plot it against anything
ggplot2::ggplot(data = EDAdata, aes(y = BodyTemp)) +
geom_boxplot()
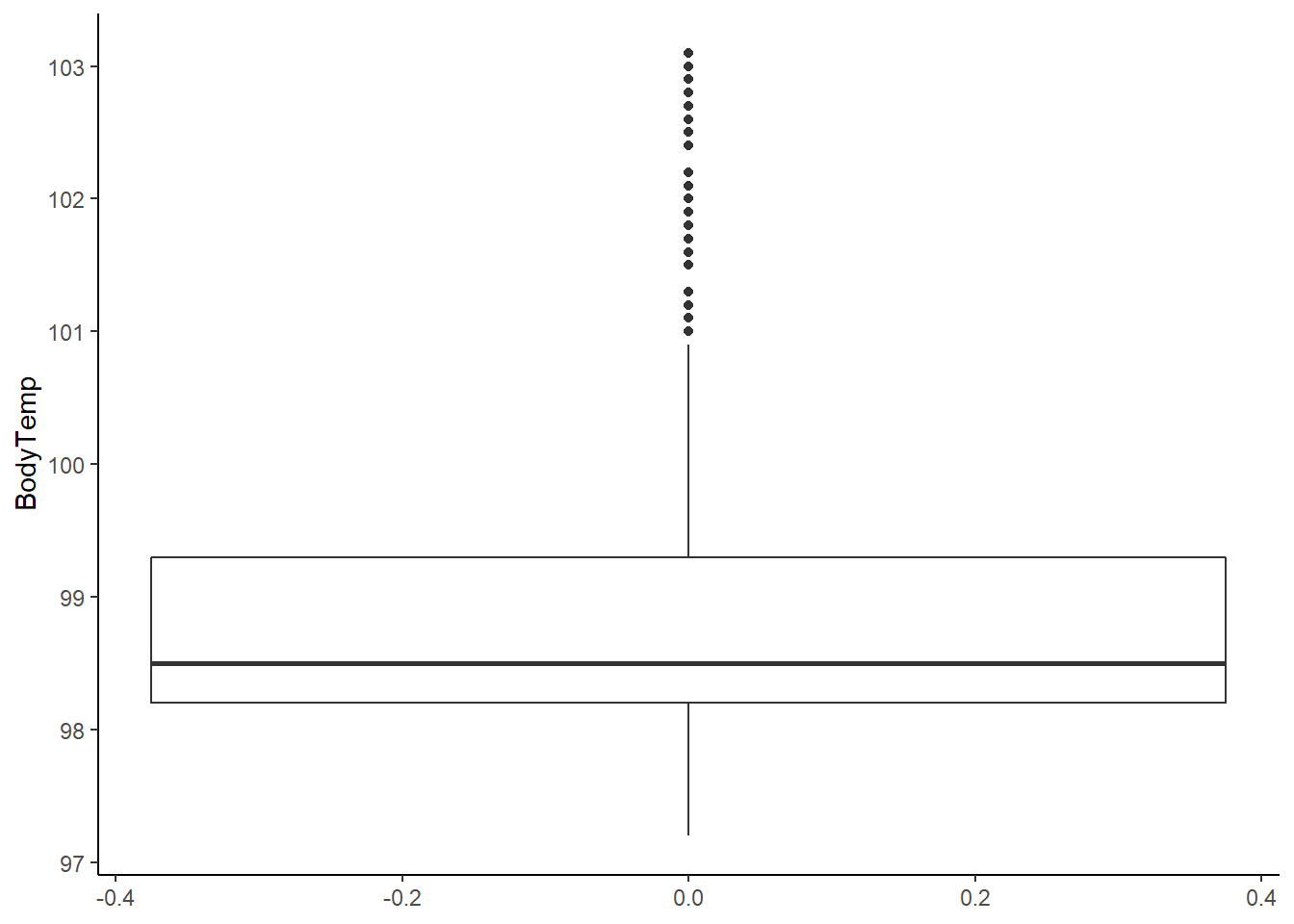
# there is clearly a skew in the data towards normal body temperature (98.6F)
# choosing to keep the points in the 101 - 103 F range as these are clinically reasonable values for influenza patients
# in other words, they are unlikely to be clinically significant outliers
# it is still worth examining the normality assumption, especially since we are moving to linear model fitting next
# (4) create a QQ-plot for body temperature
car::qqPlot(EDAdata$BodyTemp)
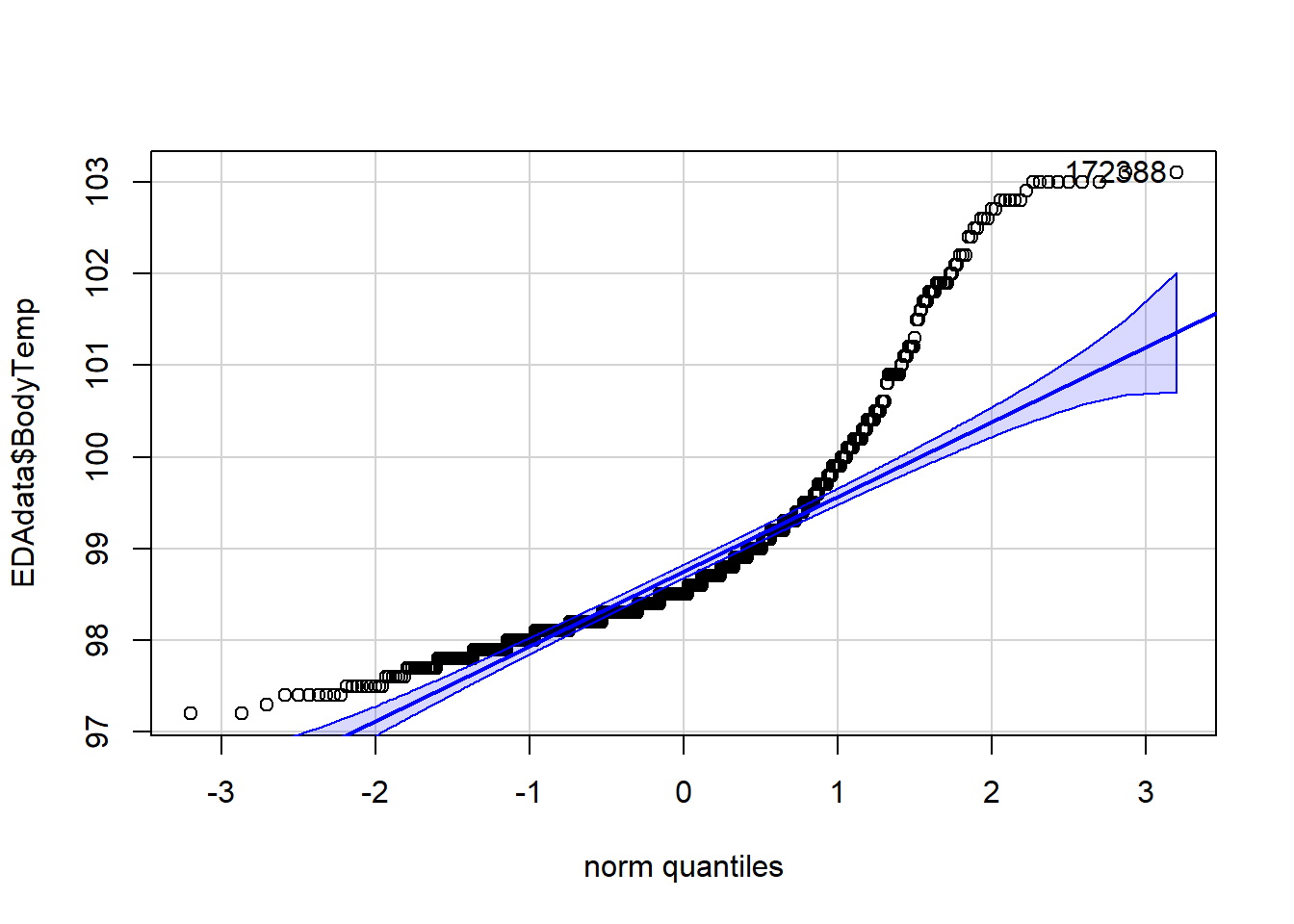
## [1] 172 388
# this clearly shows the body temperature data violates the normality assumption for linear models
Looking at the histogram and summary statistics, there appears to be a left skew from a normal distribution. The QQ plot further affirms a violation of the normality assumption. All of the values of body temperature are within clinically reasonable bounds for influenza (likely febrile) patients.
Main Categorical Outcome of Interest: Nausea
# now move onto main categorical outcome of interest: nausea
# before any analysis, create a label for nausea so outputs are more interpretable
EDAdata$Nausea <- base::factor(processeddata$Nausea, levels = c("No", "Yes"), labels = c("Nausea Absent", "Nausea Present"))
# (1) since it is categorical, we can only examine frequency and proportions of the variable
# this can be done with the summary tools package function "freq" and options to hide NAs (removed during processing)
summarytools::freq(EDAdata$Nausea, report.nas = FALSE)
## Frequencies
##
## Freq % % Cum.
## -------------------- ------ -------- --------
## Nausea Absent 475 65.07 65.07
## Nausea Present 255 34.93 100.00
## Total 730 100.00 100.00
# (2) skip as nausea is not a continuous variable
# (3) as this is a main outcome of interest, we can only create a bar plot in ggplot to illustrate distribution
# the two geom_text statements tell ggplot to calculate and display count and percentages at the top of each bar
ggplot2::ggplot(data = EDAdata, aes(x = Nausea)) +
geom_bar() +
geom_text(
aes(label = after_stat(count)),
stat = 'count',
nudge_x = -0.06,
nudge_y = 0.2,
vjust = -1) +
geom_text(
aes(label = after_stat(scales::percent(prop, prefix = "(", suffix = "%)", accuracy = 0.1)), group = 1),
stat = 'count',
nudge_x = 0.06,
nudge_y = 0.2,
vjust = -1)
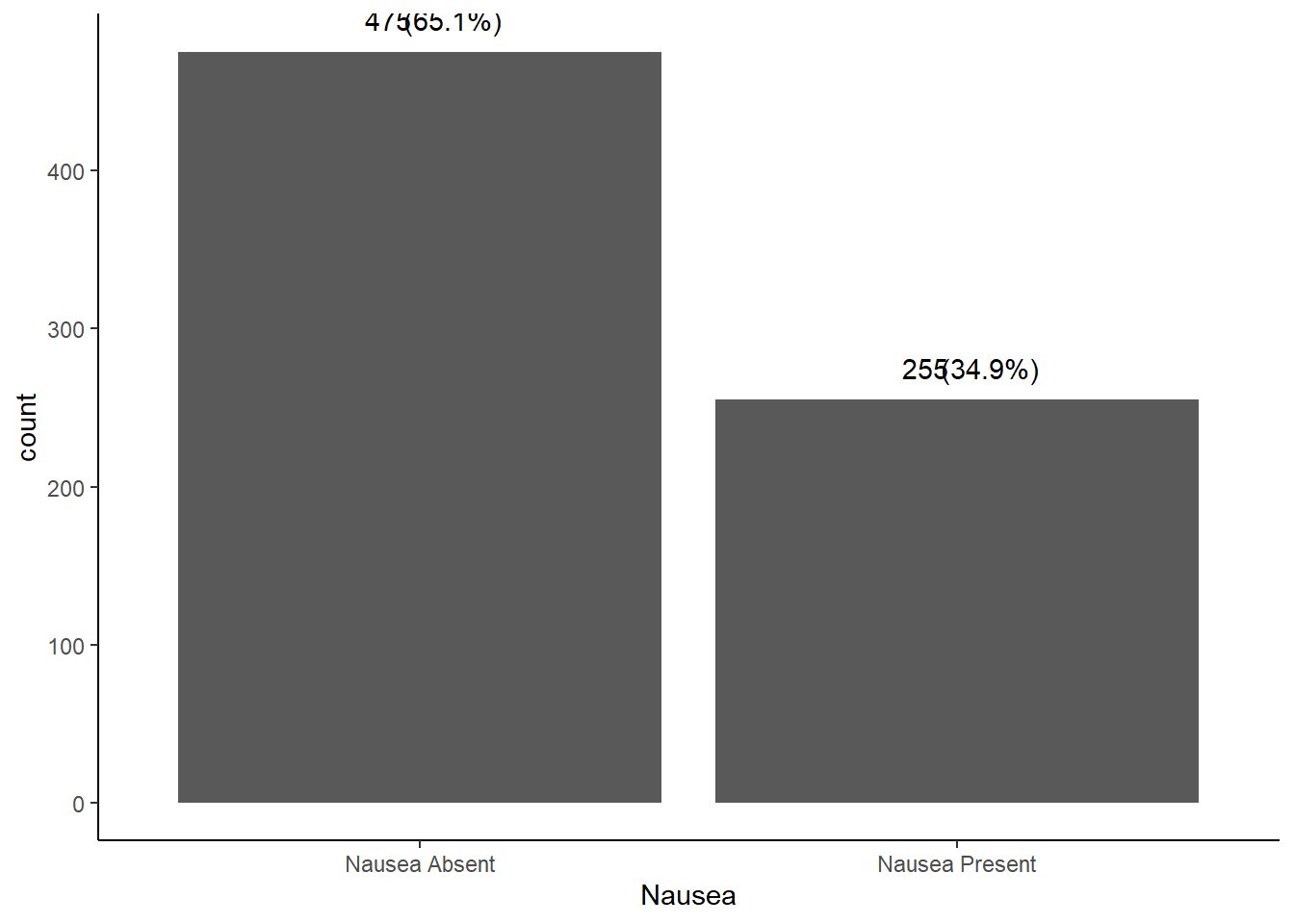
# it is almost a 2/3 vs 1/3 split for nausea in patients captured in this dataset
# there isn't much more descriptive work we can do here
The patients included in this dataset have an almost 2/3 vs 1/3 split for reports of nausea.
Predictor Variable: Runny Nose
# (1) since it is categorical, we can only examine frequency and proportions of the variable
# this can be done with the summary tools package function "freq" and options to hide NAs (removed during processing)
summarytools::freq(EDAdata$RunnyNose, report.nas = FALSE)
## Frequencies
##
## Freq % % Cum.
## ----------- ------ -------- --------
## No 211 28.90 28.90
## Yes 519 71.10 100.00
## Total 730 100.00 100.00
# almost 3/4 of the patients captured in the dataset had a runny nose
# (2) skip as runny nose is not a continuous variable
# (3) examine graphical relationship with outcomes
# start with body temperature (i.e. create a box plot)
# include a jitter function to have a better idea of number of measurements and distribution
ggplot2::ggplot(data = EDAdata, aes(x = RunnyNose, y = BodyTemp)) +
geom_boxplot() +
geom_jitter(alpha = 0.5, width = 0.2, height = 0.2, color = "tomato")
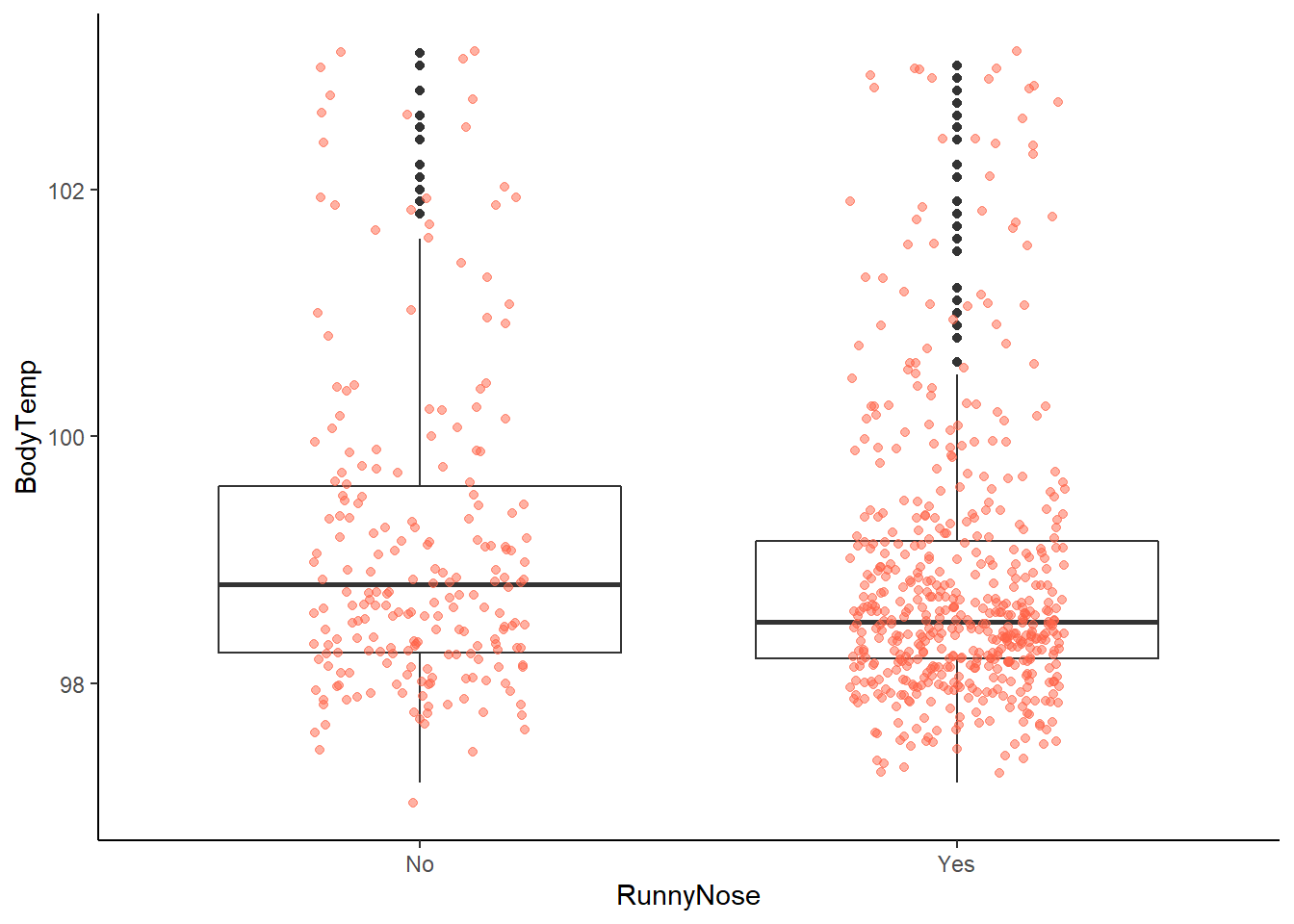
# looking at this graph, it appears that patients with runny nose are potentially less likely to have a fever
# but this requires further analysis
# (3) now with nausea
# first create a table output of runny nose by nausea
# we can do this using the table 1 package
table1::label(EDAdata$RunnyNose) <- "Runny Nose"
table1::table1(~ RunnyNose | Nausea, data = EDAdata)
|
Nausea Absent
(N=475) |
Nausea Present
(N=255) |
Overall
(N=730) |
| Runny Nose |
|
|
|
| No |
139 (29.3%) |
72 (28.2%) |
211 (28.9%) |
| Yes |
336 (70.7%) |
183 (71.8%) |
519 (71.1%) |
# since both are categorical variables, we can use a stacked bar plot to understand the distribution of runny nose within nausea symptoms
# to be able to include the percentages within each group, we will to calculate percentages before creating a graph
# first need to define a sequential piping operator so the function knows to use objects defined in the operation
`%s>%` <- magrittr::pipe_eager_lexical
# the first part of this piping operation calculates the counts and percentages within the Nausea grouping
# the second part plots it using the ggplot2 package
# trying to visualize the proportion of runny nose patients report outcome of interest (nausea)
# spacing on the labels isn't ideal, so would need to adjust for an actual manuscript
EDAdata %s>%
dplyr::group_by(RunnyNose, Nausea) %s>%
dplyr::summarise(count_Nausea = n()) %s>%
dplyr::group_by(RunnyNose) %s>%
dplyr::mutate(count_RunnyNose = sum(count_Nausea)) %s>%
dplyr::mutate(pct = count_Nausea / count_RunnyNose) %s>% {
ggplot2::ggplot(., aes(x = RunnyNose,
y = count_Nausea,
fill = Nausea)) +
ggplot2::geom_bar(
position = "stack",
stat = "identity") +
ggplot2::geom_text(
aes(label = count_Nausea),
.,
stat = 'identity',
size = 4,
position = position_stack(vjust = 0.7, reverse = FALSE)) +
ggplot2::geom_text(
aes(label = scales::percent(pct)),
.,
stat = 'identity',
size = 4,
position = position_stack(vjust = 0.5, reverse = FALSE)) +
ggplot2::labs(.,
title = "Frequency of Nausea among Runny Nose Patients",
x = "Runny Nose?",
y = "Frequency of Nausea")
}
## `summarise()` has grouped output by 'RunnyNose'. You can override using the `.groups` argument.
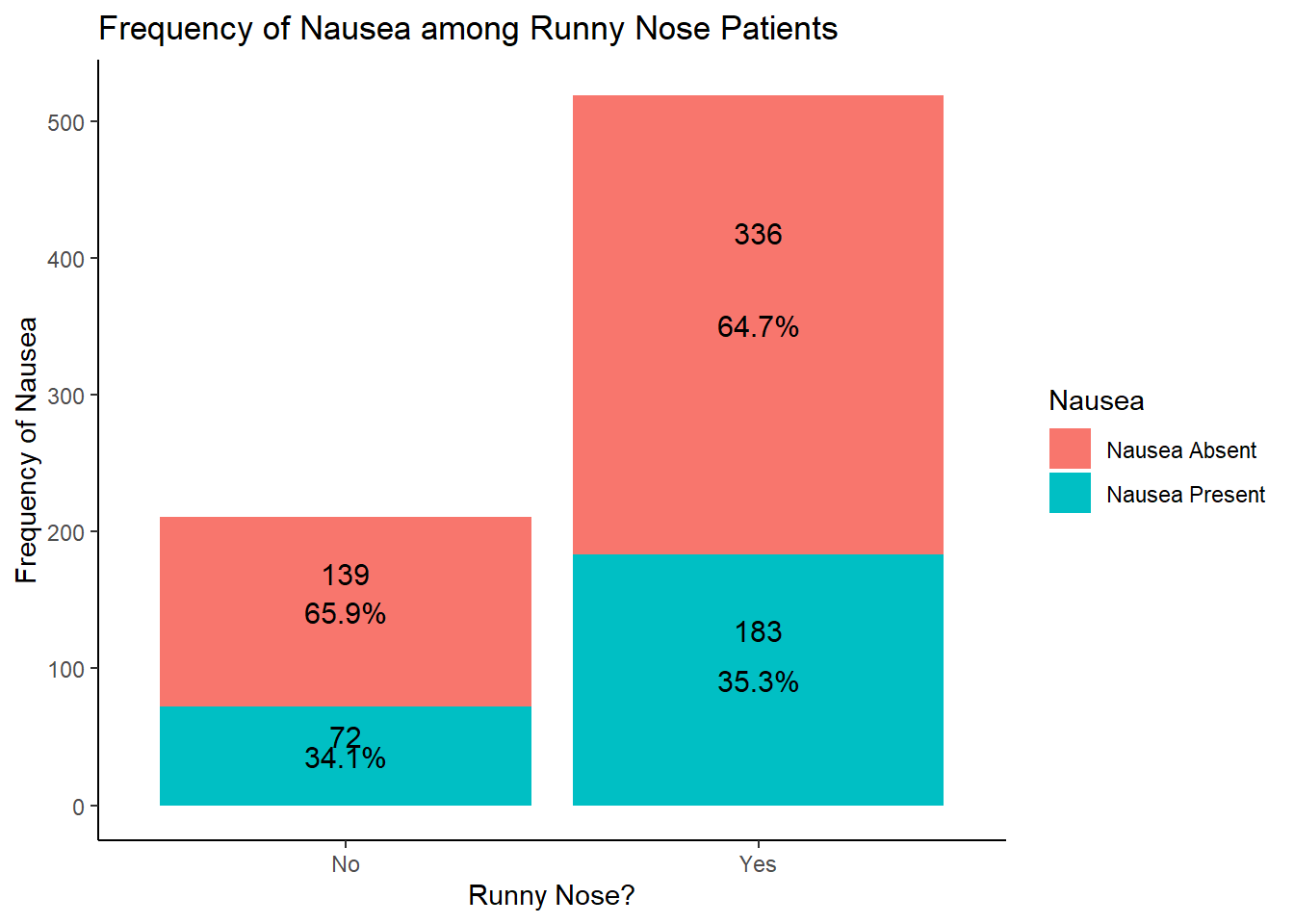
# looking at the results of this graph, it seems that the distribution of the nausea outcome isn't affected by the presence of a runny nose
Predictor Variable: Nasal Congestion
# (1) since it is categorical, we can only examine frequency and proportions of the variable
# this can be done with the summary tools package function "freq" and options to hide NAs (removed during processing)
summarytools::freq(EDAdata$NasalCongestion, report.nas = FALSE)
## Frequencies
##
## Freq % % Cum.
## ----------- ------ -------- --------
## No 167 22.88 22.88
## Yes 563 77.12 100.00
## Total 730 100.00 100.00
# more than 3/4 of the patients captured in the dataset had nasal congestion
# (2) skip as nasal congestion is not a continuous variable
# (3) examine graphical relationship with outcomes
# start with body temperature (i.e. create a box plot)
# include a jitter function to have a better idea of number of measurements and distribution
ggplot2::ggplot(data = EDAdata, aes(x = NasalCongestion, y = BodyTemp)) +
geom_boxplot() +
geom_jitter(alpha = 0.5, width = 0.2, height = 0.2, color = "tomato")
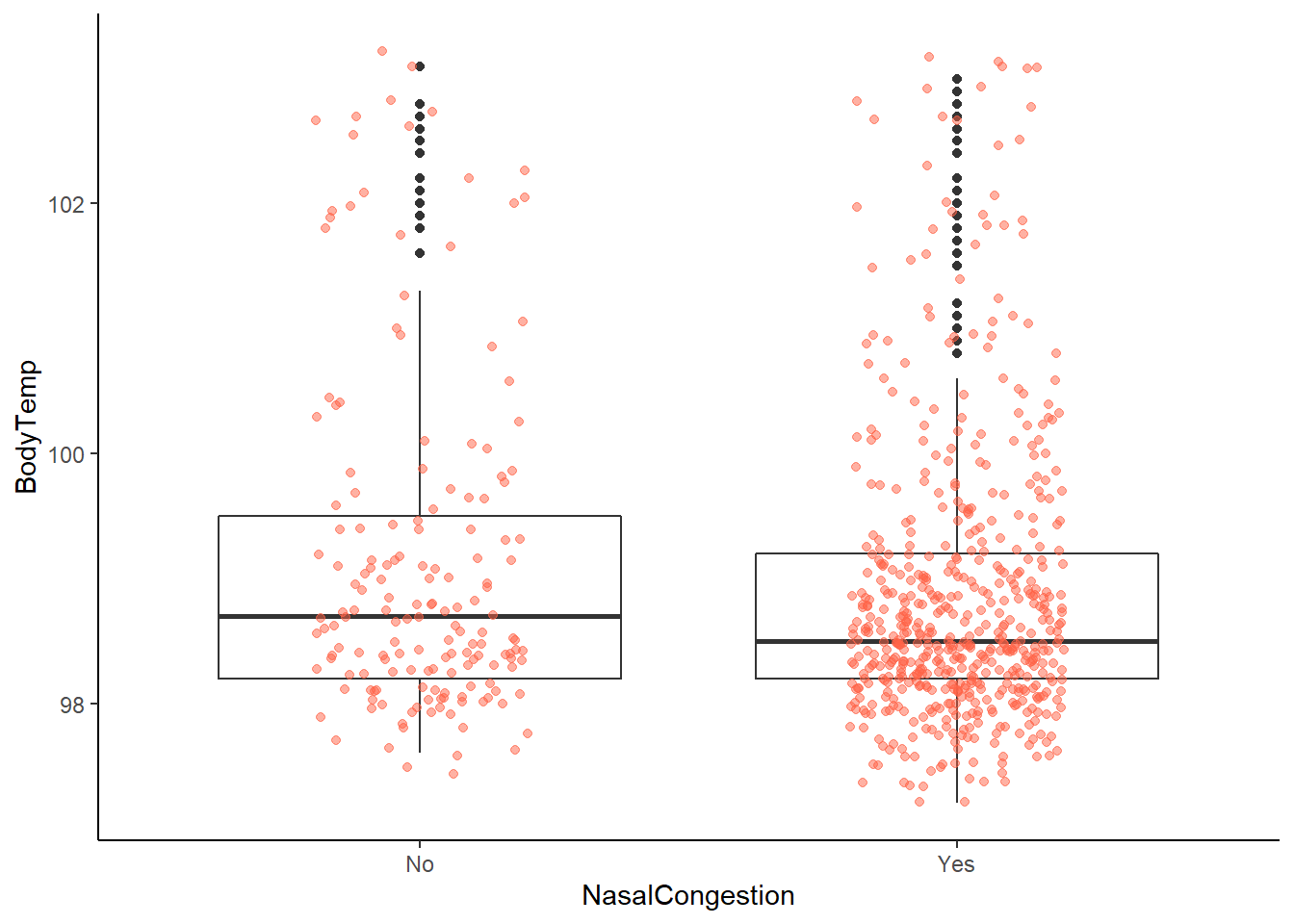
# looking at this graph, hard to tell potential difference
# likely due to totals in each group (yes = 563, no = 167)
# (3) now with nausea
# first create a table output of nasal congestion by nausea
# we can do this using the table 1 package
table1::label(EDAdata$NasalCongestion) <- "Nasal Congestion"
table1::table1(~ NasalCongestion | Nausea, data = EDAdata)
|
Nausea Absent
(N=475) |
Nausea Present
(N=255) |
Overall
(N=730) |
| Nasal Congestion |
|
|
|
| No |
120 (25.3%) |
47 (18.4%) |
167 (22.9%) |
| Yes |
355 (74.7%) |
208 (81.6%) |
563 (77.1%) |
# since both are categorical variables, we can use a stacked bar plot to understand the distribution of nausea and nasal congestion symptoms
# to be able to include the percentages within each group, we will to calculate percentages before creating a graph
# first need to define a sequential piping operator so the function knows to use objects defined in the operation
`%s>%` <- magrittr::pipe_eager_lexical
# the first part of this piping operation calculates the counts and percentages within the Nausea grouping
# the second part plots it using the ggplot2 package
# trying to visualize the proportion of nasal congestion patients report outcome of interest (nausea)
# spacing on the labels isn't ideal, so would need to adjust for an actual manuscript
EDAdata %s>%
dplyr::group_by(NasalCongestion, Nausea) %s>%
dplyr::summarise(count_Nausea = n()) %s>%
dplyr::group_by(NasalCongestion) %s>%
dplyr::mutate(count_NasalCongestion = sum(count_Nausea)) %s>%
dplyr::mutate(pct = count_Nausea / count_NasalCongestion) %s>% {
ggplot2::ggplot(., aes(x = NasalCongestion,
y = count_Nausea,
fill = Nausea)) +
ggplot2::geom_bar(
position = "stack",
stat = "identity") +
ggplot2::geom_text(
aes(label = count_Nausea),
.,
stat = 'identity',
size = 4,
position = position_stack(vjust = 0.7, reverse = FALSE)) +
ggplot2::geom_text(
aes(label = scales::percent(pct)),
.,
stat = 'identity',
size = 4,
position = position_stack(vjust = 0.5, reverse = FALSE)) +
ggplot2::labs(.,
title = "Frequency of Nausea among Nasal Congestion Patients",
x = "Nasal Congestion?",
y = "Frequency of Nausea")
}
## `summarise()` has grouped output by 'NasalCongestion'. You can override using the `.groups` argument.
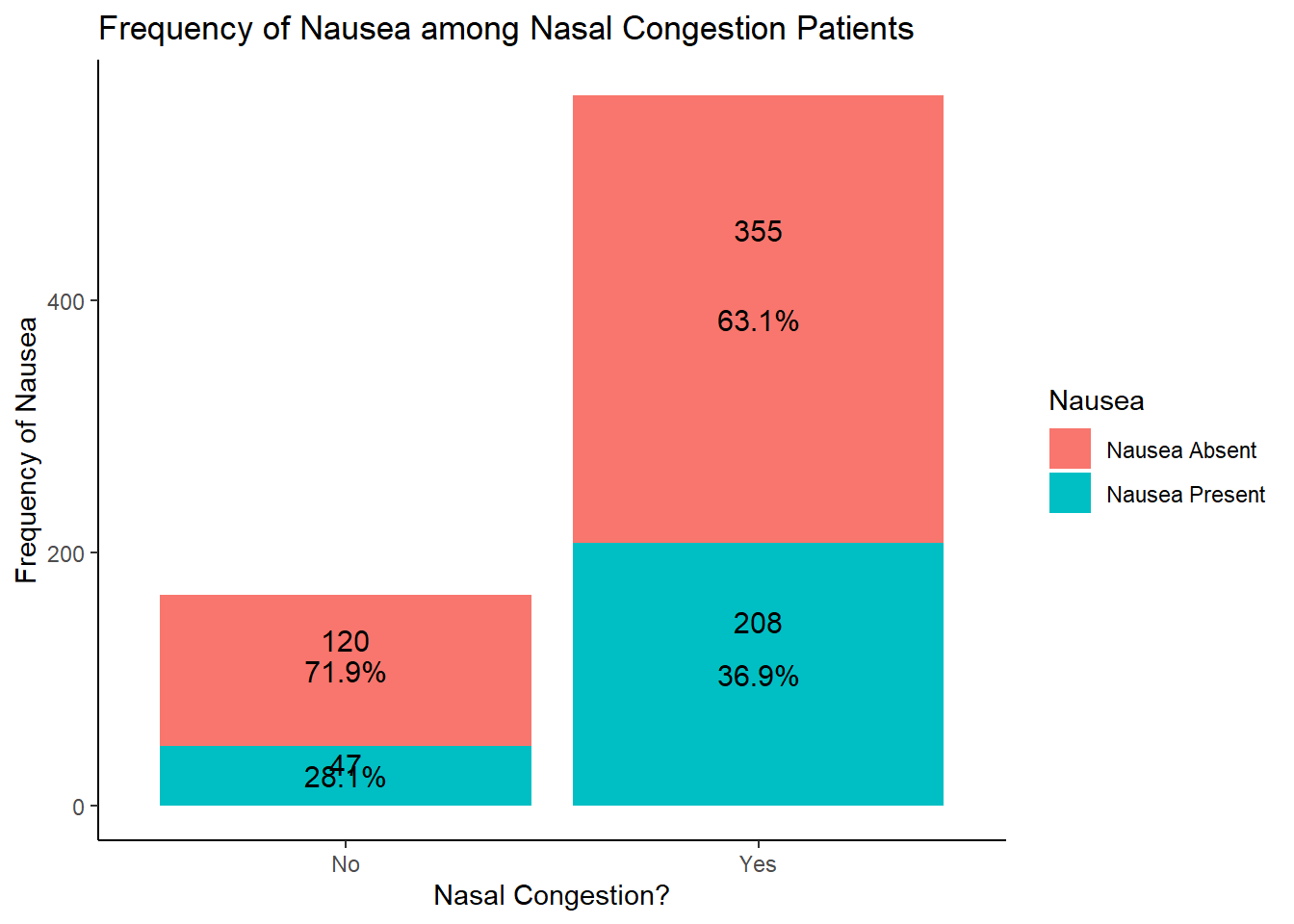
# looking at the results of this graph, potentially more likely to have nausea without nasal congestion
Predictor Variable: Pharyngitis
# (1) since it is categorical, we can only examine frequency and proportions of the variable
# this can be done with the summary tools package function "freq" and options to hide NAs (removed during processing)
summarytools::freq(EDAdata$Pharyngitis, report.nas = FALSE)
## Frequencies
##
## Freq % % Cum.
## ----------- ------ -------- --------
## No 119 16.30 16.30
## Yes 611 83.70 100.00
## Total 730 100.00 100.00
# more than 80% of patients captured in this dataset have pharyngitis
# (2) skip as phyarngitis is not a continuous variable
# (3) examine graphical relationship with outcomes
# start with body temperature (i.e. create a box plot)
# include a jitter function to have a better idea of number of measurements and distribution
ggplot2::ggplot(data = EDAdata, aes(x = Pharyngitis, y = BodyTemp)) +
geom_boxplot() +
geom_jitter(alpha = 0.5, width = 0.2, height = 0.2, color = "tomato")
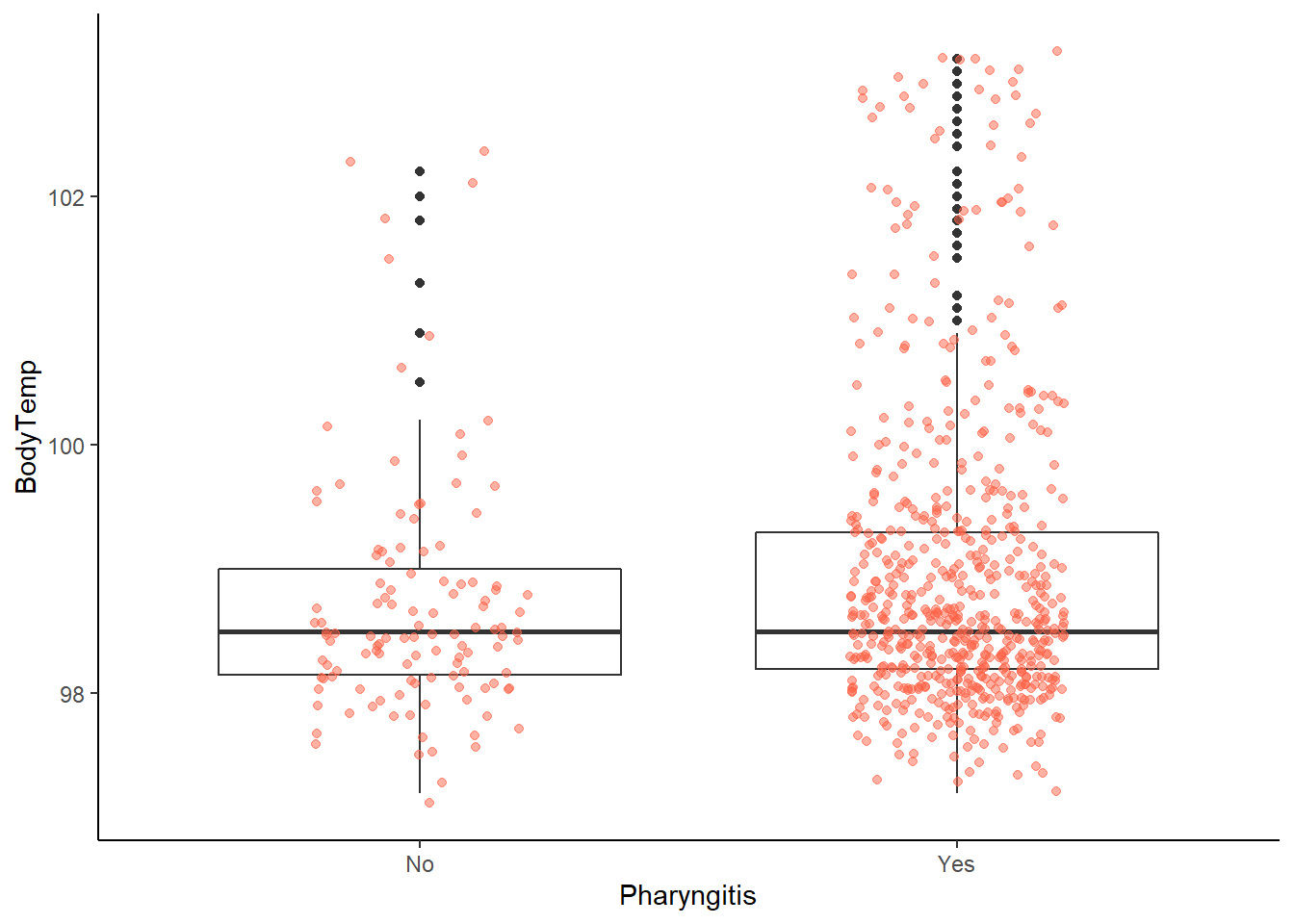
# looking at this graph, hard to tell potential difference
# likely due to totals in each group (yes = 611, no = 119)
# (3) now with nausea
# first create a table output of Pharyngitis by nausea
# we can do this using the table 1 package
table1::label(EDAdata$Pharyngitis) <- "Pharyngitis"
table1::table1(~ Pharyngitis | Nausea, data = EDAdata)
|
Nausea Absent
(N=475) |
Nausea Present
(N=255) |
Overall
(N=730) |
| Pharyngitis |
|
|
|
| No |
80 (16.8%) |
39 (15.3%) |
119 (16.3%) |
| Yes |
395 (83.2%) |
216 (84.7%) |
611 (83.7%) |
# since both are categorical variables, we can use a stacked bar plot to understand the distribution of nausea and Pharyngitis
# to be able to include the percentages within each group, we will to calculate percentages before creating a graph
# first need to define a sequential piping operator so the function knows to use objects defined in the operation
`%s>%` <- magrittr::pipe_eager_lexical
# the first part of this piping operation calculates the counts and percentages within the Nausea grouping
# the second part plots it using the ggplot2 package
# trying to visualize the proportion of Pharyngitis patients report outcome of interest (nausea)
# spacing on the labels isn't ideal, so would need to adjust for an actual manuscript
EDAdata %s>%
dplyr::group_by(Pharyngitis, Nausea) %s>%
dplyr::summarise(count_Nausea = n()) %s>%
dplyr::group_by(Pharyngitis) %s>%
dplyr::mutate(count_Pharyngitis = sum(count_Nausea)) %s>%
dplyr::mutate(pct = count_Nausea / count_Pharyngitis) %s>% {
ggplot2::ggplot(., aes(x = Pharyngitis,
y = count_Nausea,
fill = Nausea)) +
ggplot2::geom_bar(
position = "stack",
stat = "identity") +
ggplot2::geom_text(
aes(label = count_Nausea),
.,
stat = 'identity',
size = 4,
position = position_stack(vjust = 0.7, reverse = FALSE)) +
ggplot2::geom_text(
aes(label = scales::percent(pct)),
.,
stat = 'identity',
size = 4,
position = position_stack(vjust = 0.5, reverse = FALSE)) +
ggplot2::labs(.,
title = "Frequency of Nausea among Pharyngitis Patients",
x = "Pharyngitis?",
y = "Frequency of Nausea")
}
## `summarise()` has grouped output by 'Pharyngitis'. You can override using the `.groups` argument.
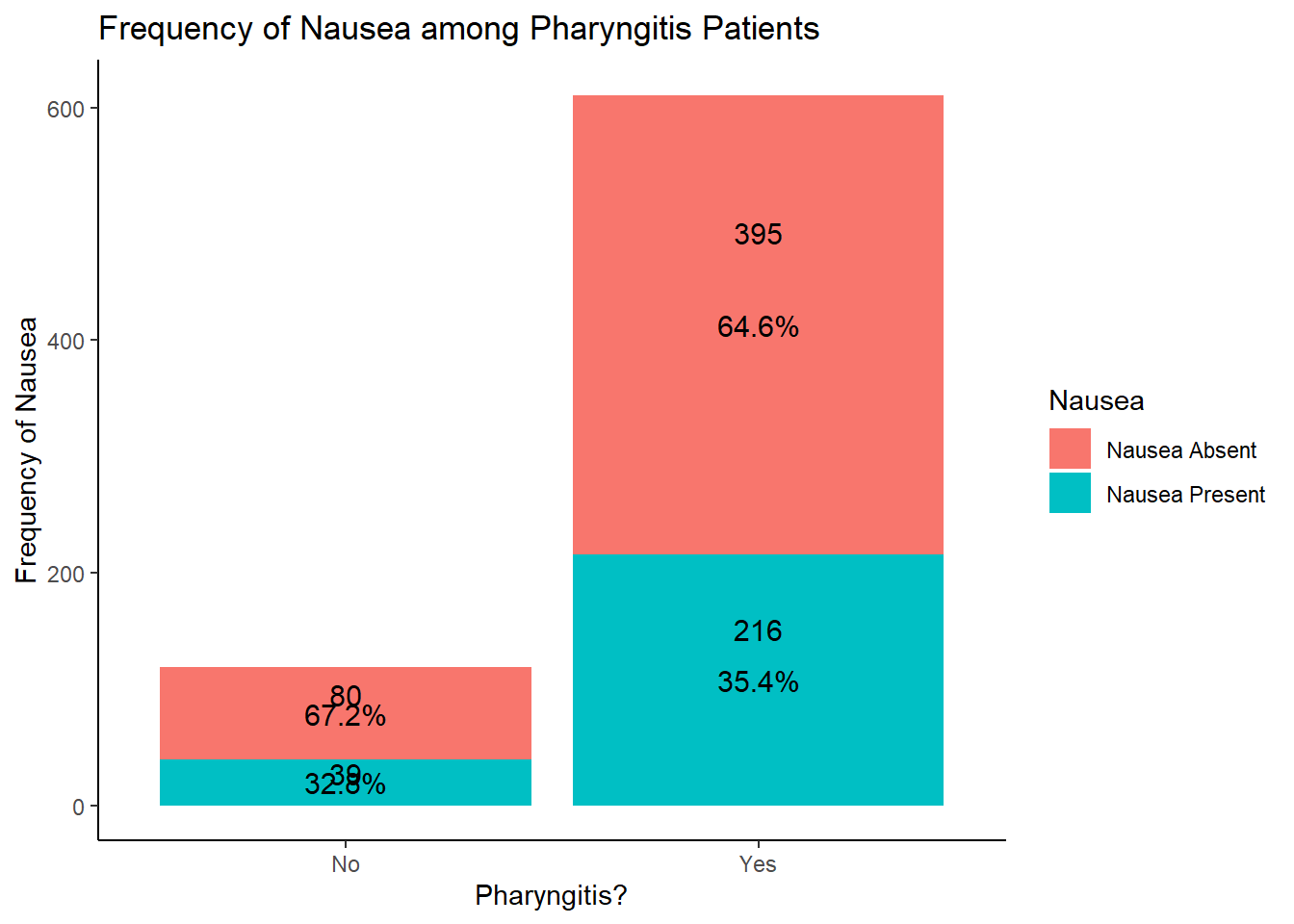
# looking at the results of this graph, hard to see any real difference
Predictor Variable: Chills / Sweating
# (1) since it is categorical, we can only examine frequency and proportions of the variable
# this can be done with the summary tools package function "freq" and options to hide NAs (removed during processing)
summarytools::freq(EDAdata$ChillsSweats, report.nas = FALSE)
## Frequencies
##
## Freq % % Cum.
## ----------- ------ -------- --------
## No 130 17.81 17.81
## Yes 600 82.19 100.00
## Total 730 100.00 100.00
# more than 80% of patients captured in this dataset have chills
# (2) skip as chills is not a continuous variable
# (3) examine graphical relationship with outcomes
# start with body temperature (i.e. create a box plot)
# include a jitter function to have a better idea of number of measurements and distribution
ggplot2::ggplot(data = EDAdata, aes(x = ChillsSweats, y = BodyTemp)) +
geom_boxplot() +
geom_jitter(alpha = 0.5, width = 0.2, height = 0.2, color = "tomato")
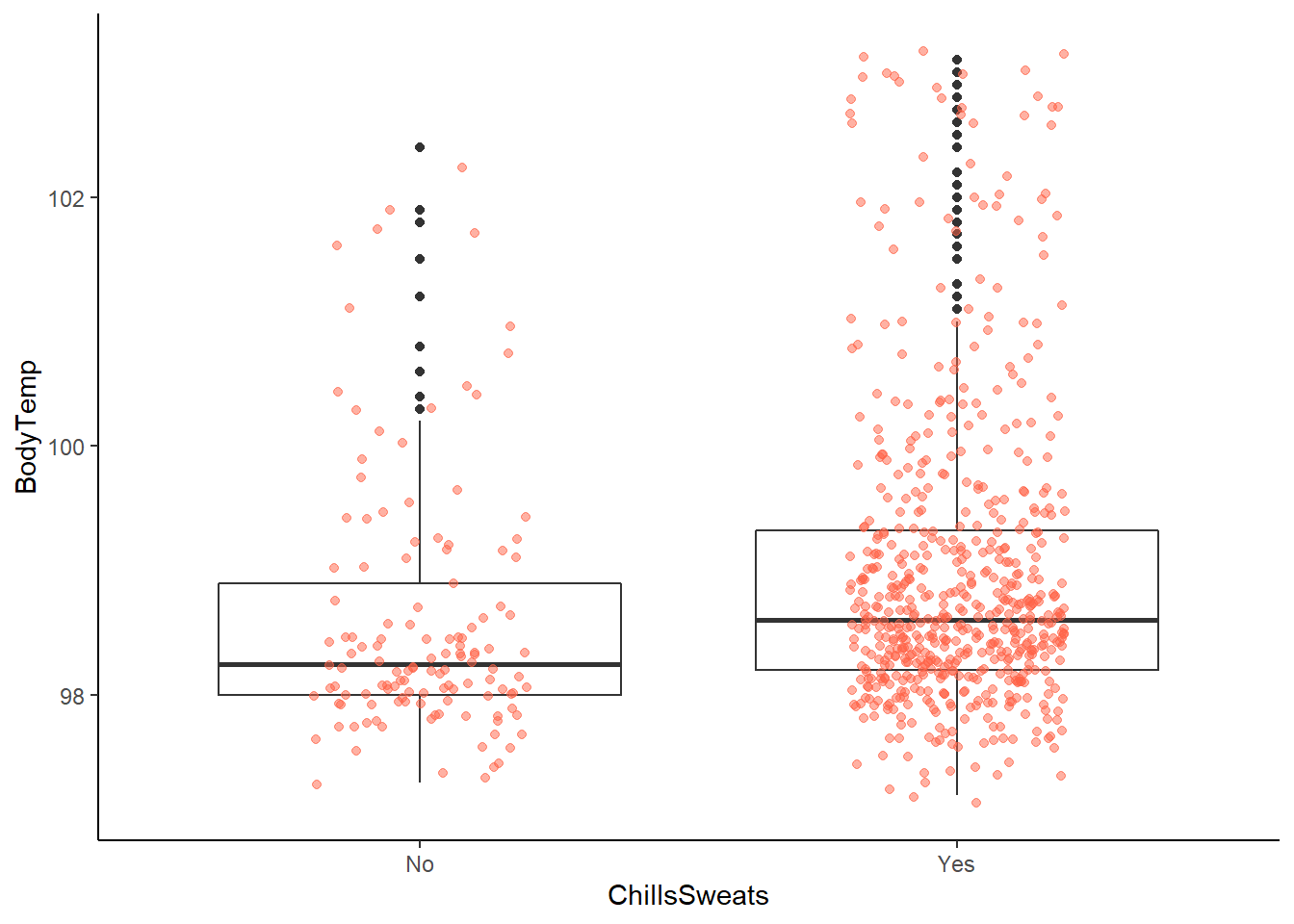
# more chills / sweats reported with higher body temperature
# this difference makes sense as chills / sweats are often the result of a fever
# (3) now with nausea
# first create a table output of chills by nausea
# we can do this using the table 1 package
table1::label(EDAdata$ChillsSweats) <- "ChillsSweats"
table1::table1(~ ChillsSweats | Nausea, data = EDAdata)
|
Nausea Absent
(N=475) |
Nausea Present
(N=255) |
Overall
(N=730) |
| ChillsSweats |
|
|
|
| No |
103 (21.7%) |
27 (10.6%) |
130 (17.8%) |
| Yes |
372 (78.3%) |
228 (89.4%) |
600 (82.2%) |
# since both are categorical variables, we can use a stacked bar plot to understand the distribution of nausea and chills
# to be able to include the percentages within each group, we will to calculate percentages before creating a graph
# first need to define a sequential piping operator so the function knows to use objects defined in the operation
`%s>%` <- magrittr::pipe_eager_lexical
# the first part of this piping operation calculates the counts and percentages within the Nausea grouping
# the second part plots it using the ggplot2 package
# trying to visualize the proportion of chills patients report outcome of interest (nausea)
# spacing on the labels isn't ideal, so would need to adjust for an actual manuscript
EDAdata %s>%
dplyr::group_by(ChillsSweats, Nausea) %s>%
dplyr::summarise(count_Nausea = n()) %s>%
dplyr::group_by(ChillsSweats) %s>%
dplyr::mutate(count_ChillsSweats = sum(count_Nausea)) %s>%
dplyr::mutate(pct = count_Nausea / count_ChillsSweats) %s>% {
ggplot2::ggplot(., aes(x = ChillsSweats,
y = count_Nausea,
fill = Nausea)) +
ggplot2::geom_bar(
position = "stack",
stat = "identity") +
ggplot2::geom_text(
aes(label = count_Nausea),
.,
stat = 'identity',
size = 4,
position = position_stack(vjust = 0.7, reverse = FALSE)) +
ggplot2::geom_text(
aes(label = scales::percent(pct)),
.,
stat = 'identity',
size = 4,
position = position_stack(vjust = 0.5, reverse = FALSE)) +
ggplot2::labs(.,
title = "Frequency of Nausea among Chills / Sweats",
x = "Chills or Sweats?",
y = "Frequency of Nausea")
}
## `summarise()` has grouped output by 'ChillsSweats'. You can override using the `.groups` argument.
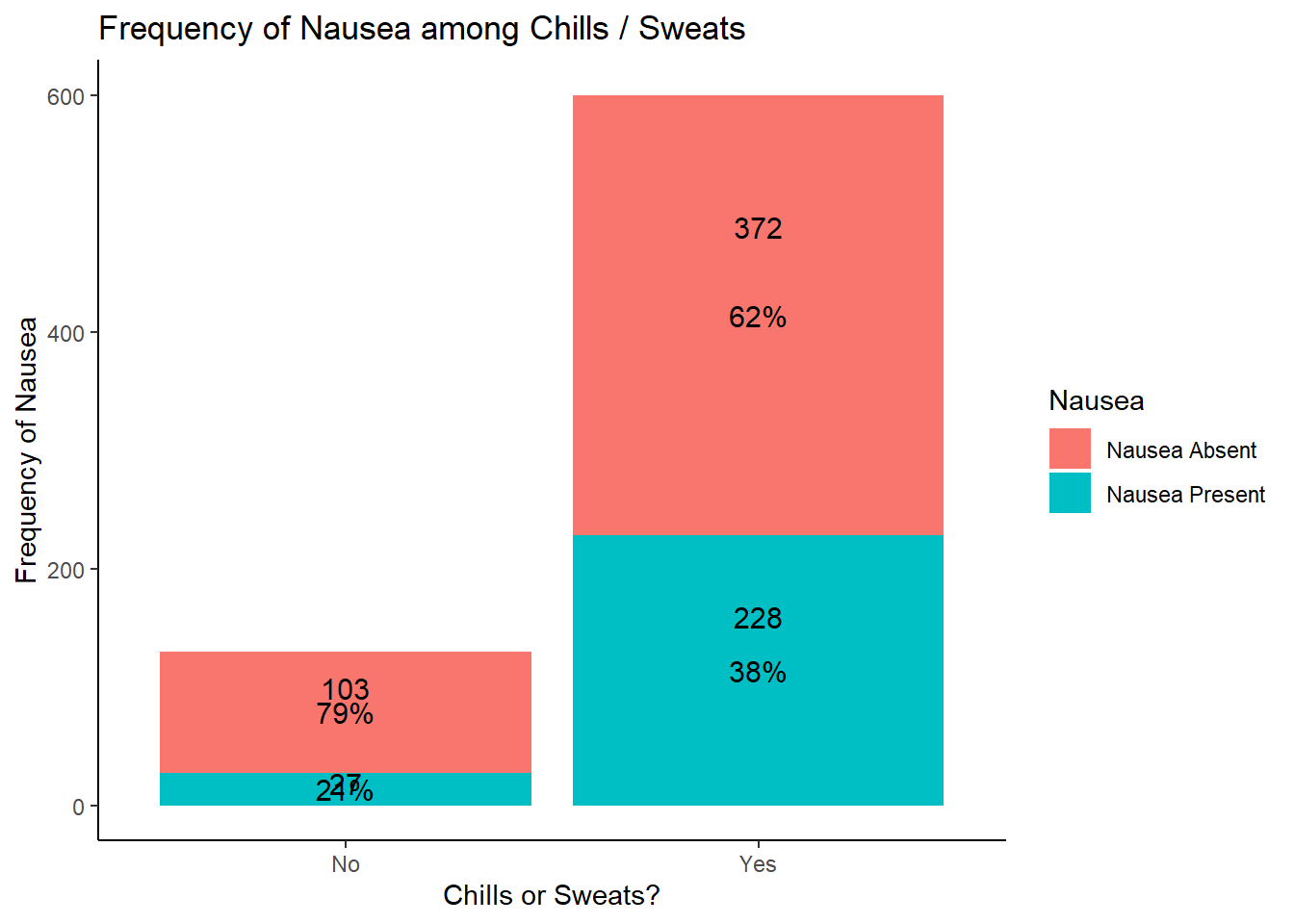
# looking at the results of this graph, potentially more nausea with chills / sweats
# requires further analysis to determine significance
Predictor Variable: Myalgia
# there are multiple variables for myalgia, but we can focus on the one that gives a severity scale of myalgia
# (1) since it is categorical, we can only examine frequency and proportions of the variable
# this can be done with the summary tools package function "freq" and options to hide NAs (removed during processing)
summarytools::freq(EDAdata$Myalgia, report.nas = FALSE)
## Frequencies
##
## Freq % % Cum.
## -------------- ------ -------- --------
## None 79 10.82 10.82
## Mild 213 29.18 40.00
## Moderate 325 44.52 84.52
## Severe 113 15.48 100.00
## Total 730 100.00 100.00
# nearly half of the patients in the dataset reported moderate myalgia
# approximately 3/4 of the patients in the dataset reported mild or moderate myalgia
# (2) skip as myalgia is not a continuous variable
# (3) examine graphical relationship with outcomes
# start with body temperature (i.e. create a box plot)
# include a jitter function to have a better idea of number of measurements and distribution
ggplot2::ggplot(data = EDAdata, aes(x = Myalgia, y = BodyTemp)) +
geom_boxplot() +
geom_jitter(alpha = 0.5, width = 0.2, height = 0.2, color = "tomato")
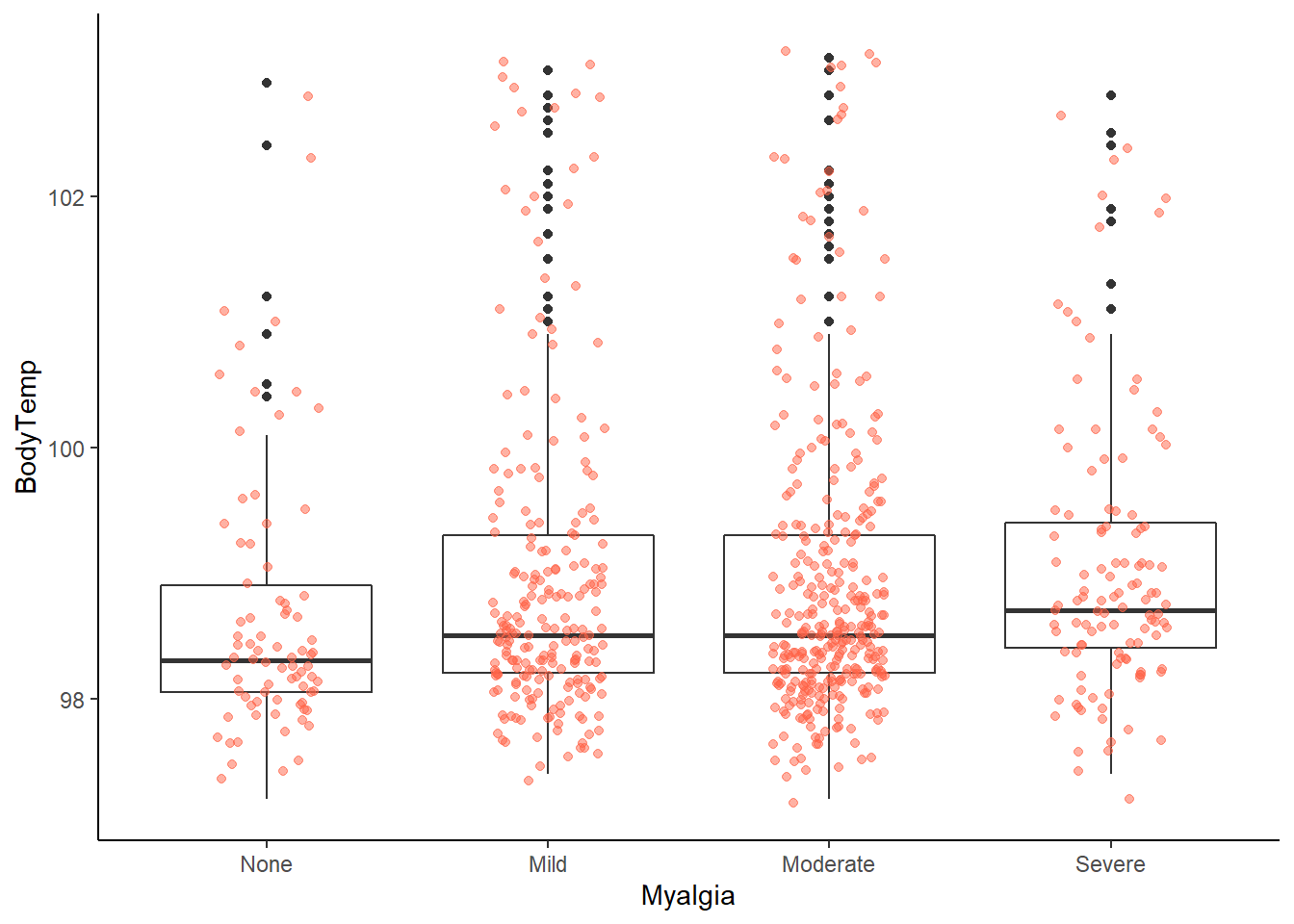
# looking at this graph, it appears that patients with no myalgia were less likely to have a fever
# it doesn't appear to have a great variation among the severity of myalgia symptoms
# but this requires further analysis
# (3) now with nausea
# first create a table output of runny nose by nausea
# we can do this using the table 1 package
table1::label(EDAdata$Myalgia) <- "Myalgia"
table1::table1(~ Myalgia | Nausea, data = EDAdata)
|
Nausea Absent
(N=475) |
Nausea Present
(N=255) |
Overall
(N=730) |
| Myalgia |
|
|
|
| None |
63 (13.3%) |
16 (6.3%) |
79 (10.8%) |
| Mild |
159 (33.5%) |
54 (21.2%) |
213 (29.2%) |
| Moderate |
198 (41.7%) |
127 (49.8%) |
325 (44.5%) |
| Severe |
55 (11.6%) |
58 (22.7%) |
113 (15.5%) |
# since both are categorical variables, we can use a stacked bar plot to understand the distribution of nausea within myalgia symptoms
# to be able to include the percentages within each group, we will to calculate percentages before creating a graph
# first need to define a sequential piping operator so the function knows to use objects defined in the operation
`%s>%` <- magrittr::pipe_eager_lexical
# the first part of this piping operation calculates the counts and percentages within the Nausea grouping
# the second part plots it using the ggplot2 package
# trying to visualize the proportion of myalgia patients report outcome of interest (nausea)
# spacing on the labels isn't ideal, so would need to adjust for an actual manuscript
EDAdata %s>%
dplyr::group_by(Myalgia, Nausea) %s>%
dplyr::summarise(count_Nausea = n()) %s>%
dplyr::group_by(Myalgia) %s>%
dplyr::mutate(count_Myalgia = sum(count_Nausea)) %s>%
dplyr::mutate(pct = count_Nausea / count_Myalgia) %s>% {
ggplot2::ggplot(., aes(x = Myalgia,
y = count_Nausea,
fill = Nausea)) +
ggplot2::geom_bar(
position = "stack",
stat = "identity") +
ggplot2::geom_text(
aes(label = count_Nausea),
.,
stat = 'identity',
size = 4,
position = position_stack(vjust = 0.7, reverse = FALSE)) +
ggplot2::geom_text(
aes(label = scales::percent(pct)),
.,
stat = 'identity',
size = 4,
position = position_stack(vjust = 0.5, reverse = FALSE)) +
ggplot2::labs(.,
title = "Frequency of Nausea among Myalgia Severity",
x = "Myalgia Severity",
y = "Frequency of Nausea")
}
## `summarise()` has grouped output by 'Myalgia'. You can override using the `.groups` argument.
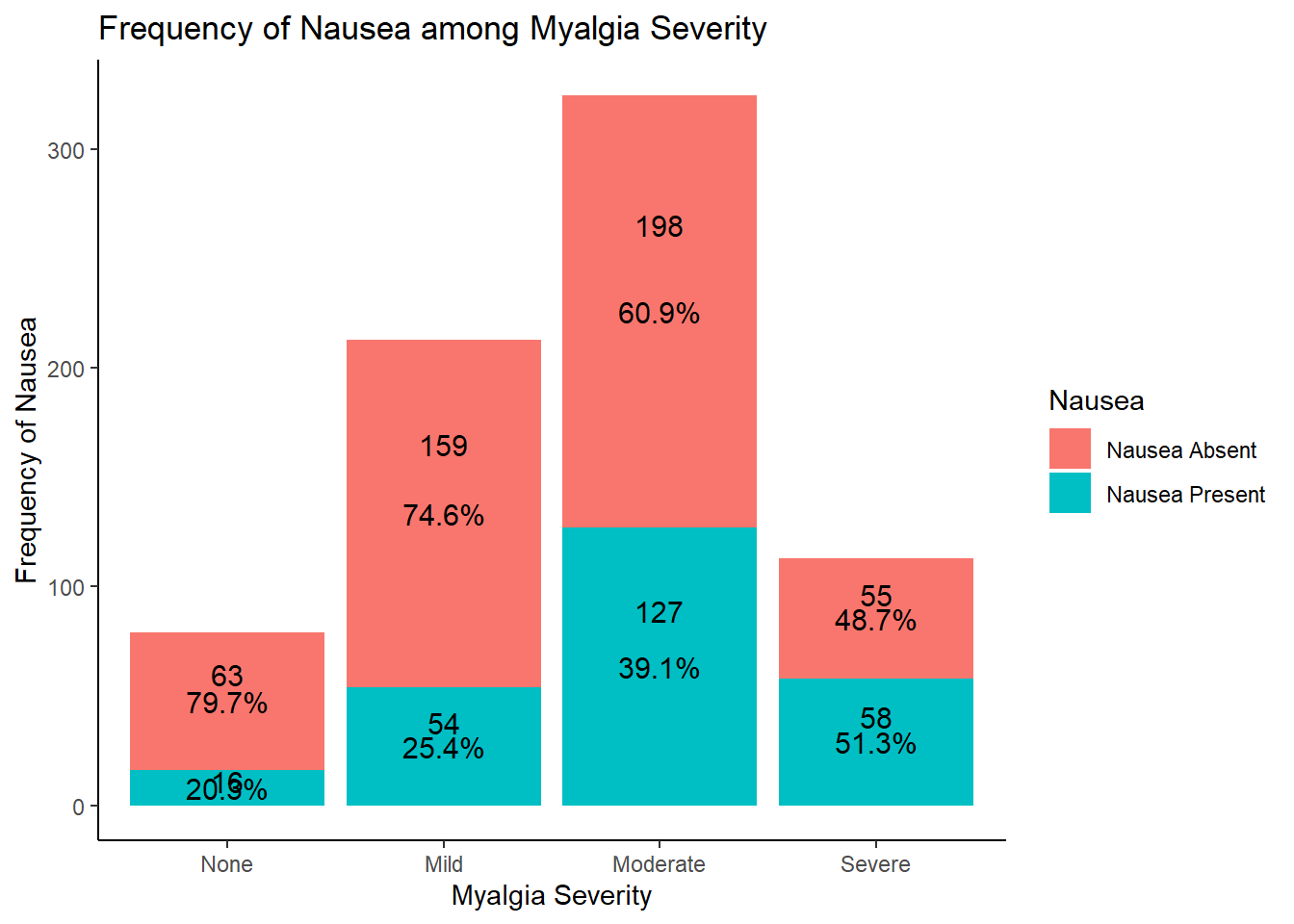
# looking at the results of this graph, it seems that increasing myalgia severity is associated with decreased nausea
# this makes sense clinically
# need further evaluation for significance
Predictor Variable: Diarrhea
# (1) since it is categorical, we can only examine frequency and proportions of the variable
# this can be done with the summary tools package function "freq" and options to hide NAs (removed during processing)
summarytools::freq(EDAdata$Diarrhea, report.nas = FALSE)
## Frequencies
##
## Freq % % Cum.
## ----------- ------ -------- --------
## No 631 86.44 86.44
## Yes 99 13.56 100.00
## Total 730 100.00 100.00
# more than 80% of patients captured in this dataset have Diarrhea
# (2) skip as Diarrhea is not a continuous variable
# (3) examine graphical relationship with outcomes
# start with body temperature (i.e. create a box plot)
# include a jitter function to have a better idea of number of measurements and distribution
ggplot2::ggplot(data = EDAdata, aes(x = Diarrhea, y = BodyTemp)) +
geom_boxplot() +
geom_jitter(alpha = 0.5, width = 0.2, height = 0.2, color = "tomato")
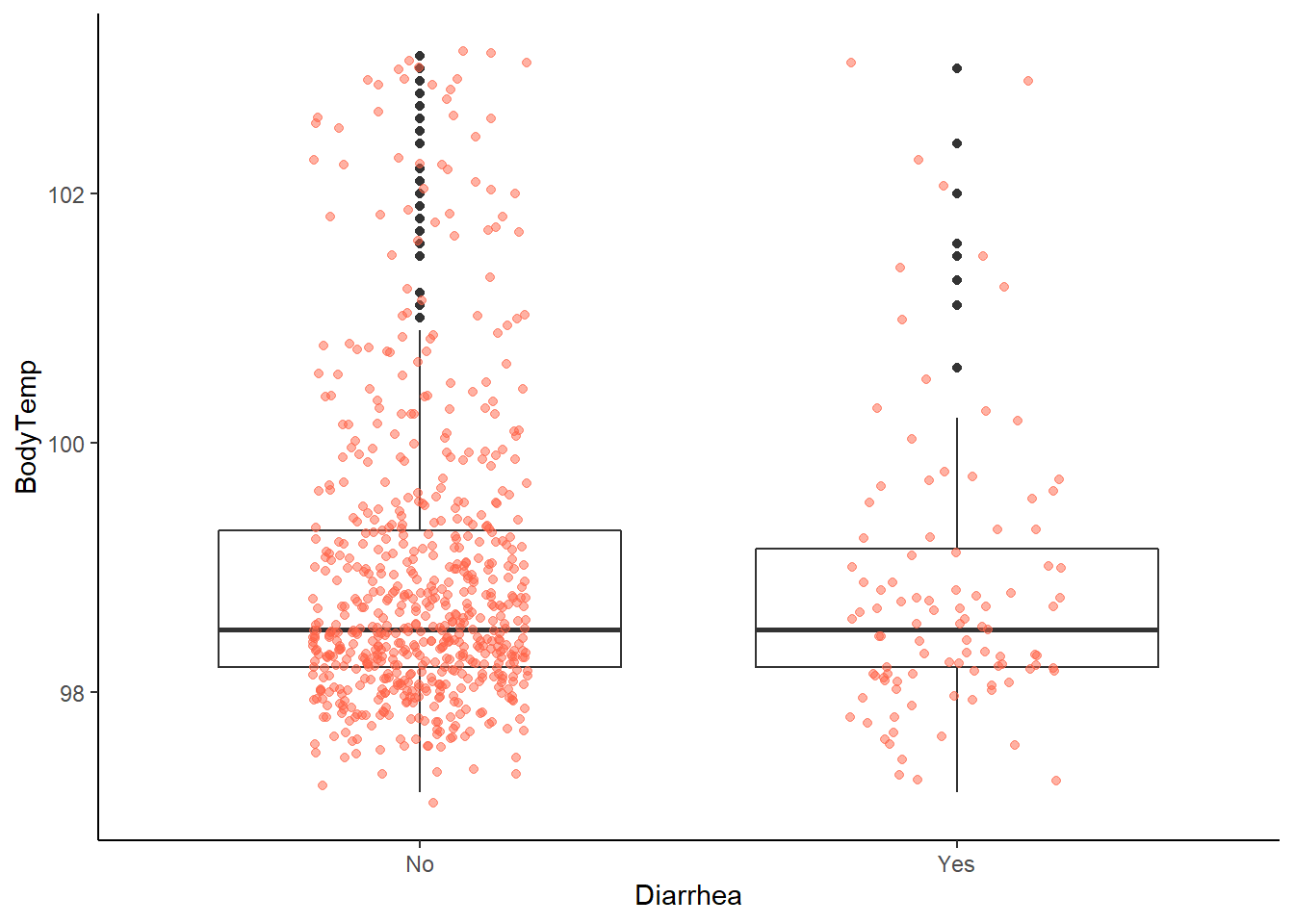
# no clear difference in body temperature
# (3) now with nausea
# first create a table output of Diarrhea by nausea
# we can do this using the table 1 package
table1::label(EDAdata$Diarrhea) <- "Diarrhea"
table1::table1(~ Diarrhea | Nausea, data = EDAdata)
|
Nausea Absent
(N=475) |
Nausea Present
(N=255) |
Overall
(N=730) |
| Diarrhea |
|
|
|
| No |
435 (91.6%) |
196 (76.9%) |
631 (86.4%) |
| Yes |
40 (8.4%) |
59 (23.1%) |
99 (13.6%) |
# since both are categorical variables, we can use a stacked bar plot to understand the distribution of nausea and Diarrhea
# to be able to include the percentages within each group, we will to calculate percentages before creating a graph
# first need to define a sequential piping operator so the function knows to use objects defined in the operation
`%s>%` <- magrittr::pipe_eager_lexical
# the first part of this piping operation calculates the counts and percentages within the Nausea grouping
# the second part plots it using the ggplot2 package
# trying to visualize the proportion of Diarrhea patients report outcome of interest (nausea)
# spacing on the labels isn't ideal, so would need to adjust for an actual manuscript
EDAdata %s>%
dplyr::group_by(Diarrhea, Nausea) %s>%
dplyr::summarise(count_Nausea = n()) %s>%
dplyr::group_by(Diarrhea) %s>%
dplyr::mutate(count_Diarrhea = sum(count_Nausea)) %s>%
dplyr::mutate(pct = count_Nausea / count_Diarrhea) %s>% {
ggplot2::ggplot(., aes(x = Diarrhea,
y = count_Nausea,
fill = Nausea)) +
ggplot2::geom_bar(
position = "stack",
stat = "identity") +
ggplot2::geom_text(
aes(label = count_Nausea),
.,
stat = 'identity',
size = 4,
position = position_stack(vjust = 0.7, reverse = FALSE)) +
ggplot2::geom_text(
aes(label = scales::percent(pct)),
.,
stat = 'identity',
size = 4,
position = position_stack(vjust = 0.5, reverse = FALSE)) +
ggplot2::labs(.,
title = "Frequency of Nausea among Diarrhea",
x = "Diarrhea?",
y = "Frequency of Nausea")
}
## `summarise()` has grouped output by 'Diarrhea'. You can override using the `.groups` argument.
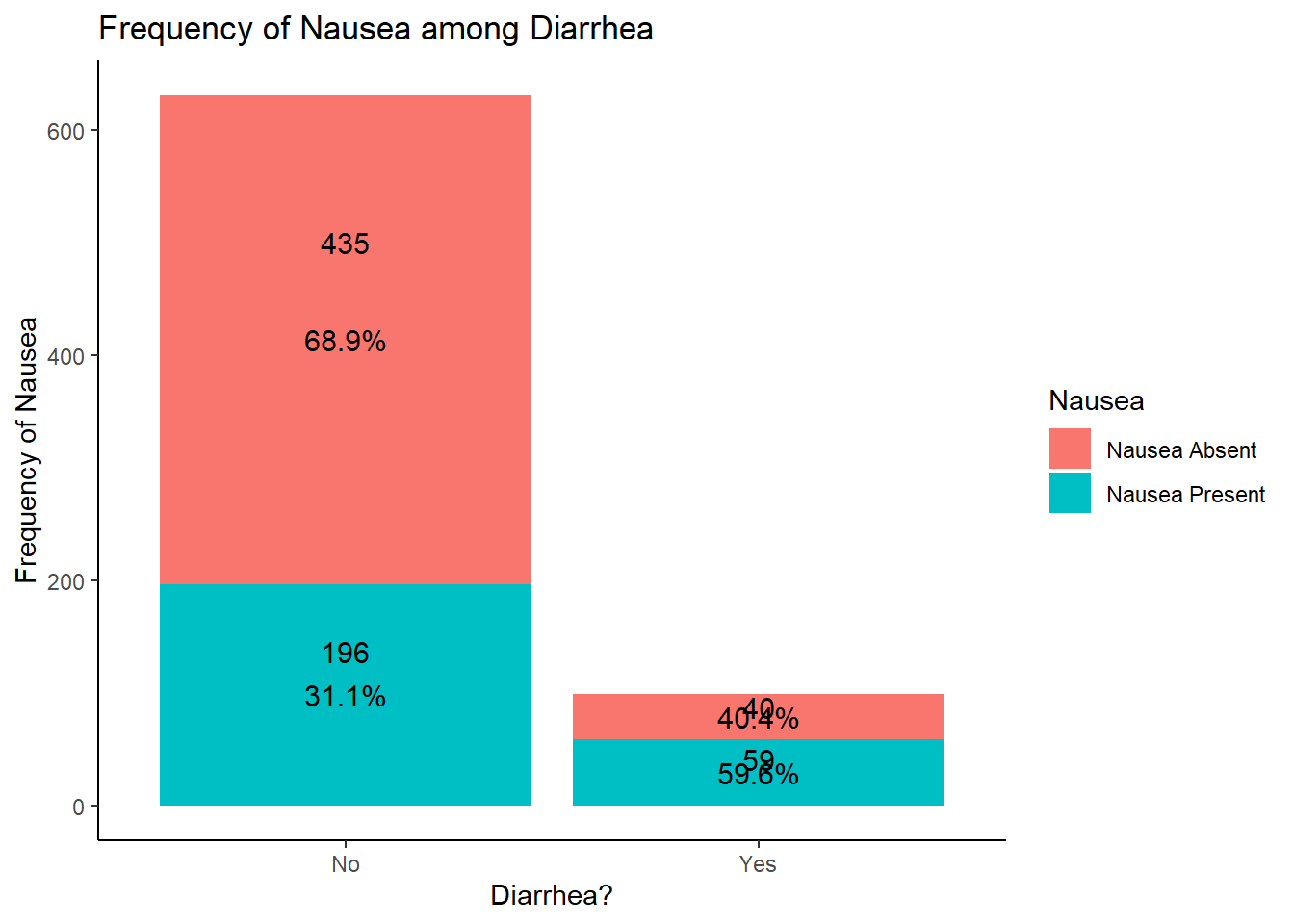
# based on the results, it appears that more patients with diarrhea had nausea
# this makes sense clinically as nausea and diarrhea often co-present
Predictor Variable: Vomitting
# (1) since it is categorical, we can only examine frequency and proportions of the variable
# this can be done with the summary tools package function "freq" and options to hide NAs (removed during processing)
summarytools::freq(EDAdata$Vomit, report.nas = FALSE)
## Frequencies
##
## Freq % % Cum.
## ----------- ------ -------- --------
## No 652 89.32 89.32
## Yes 78 10.68 100.00
## Total 730 100.00 100.00
# more than 80% of patients captured in this dataset report vomiting
# (2) skip as Vomit is not a continuous variable
# (3) examine graphical relationship with outcomes
# start with body temperature (i.e. create a box plot)
# include a jitter function to have a better idea of number of measurements and distribution
ggplot2::ggplot(data = EDAdata, aes(x = Vomit, y = BodyTemp)) +
geom_boxplot() +
geom_jitter(alpha = 0.5, width = 0.2, height = 0.2, color = "tomato")
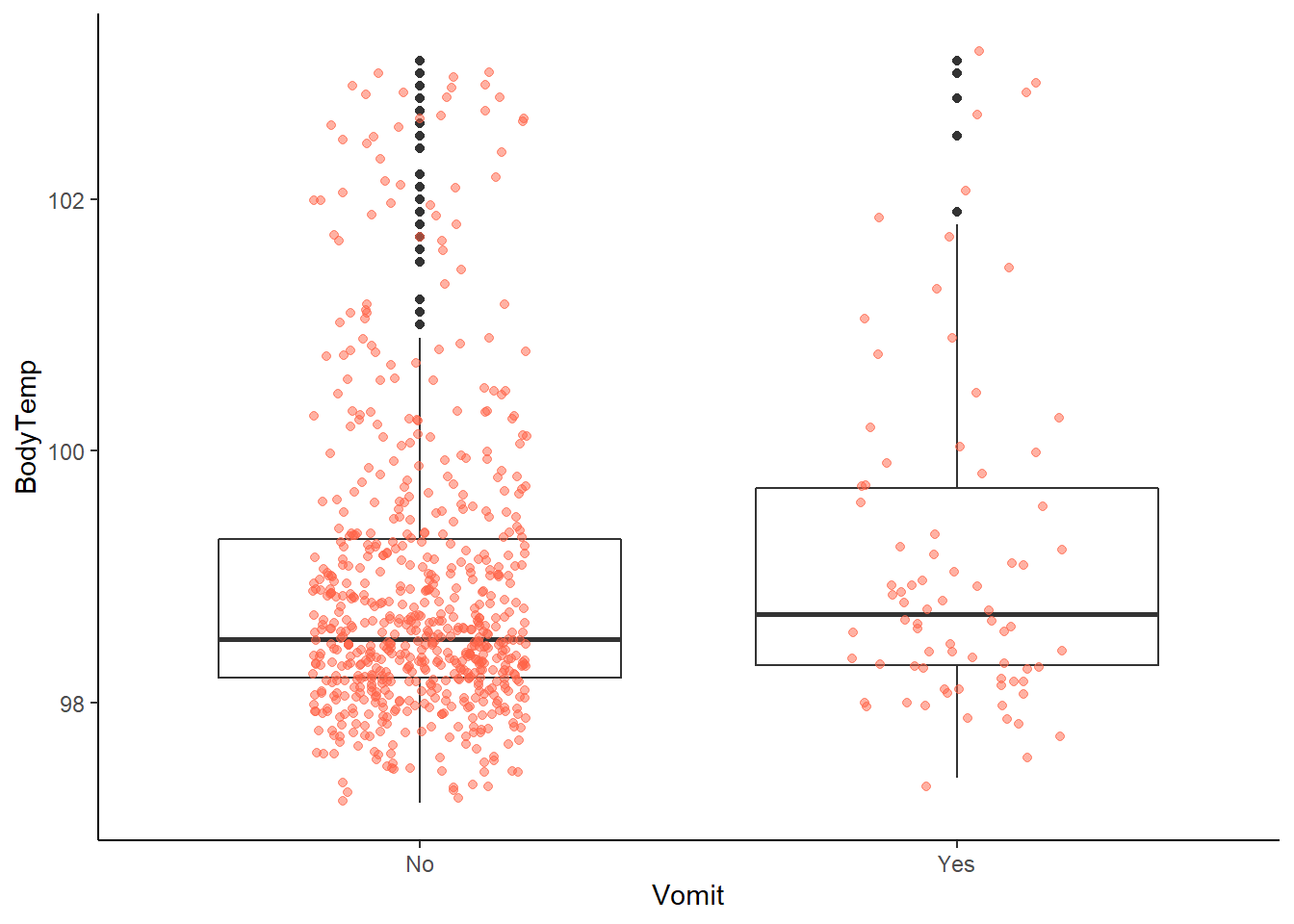
# most obviously far fewer patients reporting vomiting
# but, potentially associated with an increased body temperature
# (3) now with nausea
# first create a table output of Vomit by nausea
# we can do this using the table 1 package
table1::label(EDAdata$Vomit) <- "Vomit"
table1::table1(~ Vomit | Nausea, data = EDAdata)
|
Nausea Absent
(N=475) |
Nausea Present
(N=255) |
Overall
(N=730) |
| Vomit |
|
|
|
| No |
461 (97.1%) |
191 (74.9%) |
652 (89.3%) |
| Yes |
14 (2.9%) |
64 (25.1%) |
78 (10.7%) |
# since both are categorical variables, we can use a stacked bar plot to understand the distribution of nausea and vomiting
# to be able to include the percentages within each group, we will to calculate percentages before creating a graph
# first need to define a sequential piping operator so the function knows to use objects defined in the operation
`%s>%` <- magrittr::pipe_eager_lexical
# the first part of this piping operation calculates the counts and percentages within the Nausea grouping
# the second part plots it using the ggplot2 package
# trying to visualize the proportion of vomiting patients report outcome of interest (nausea)
# spacing on the labels isn't ideal, so would need to adjust for an actual manuscript
EDAdata %s>%
dplyr::group_by(Vomit, Nausea) %s>%
dplyr::summarise(count_Nausea = n()) %s>%
dplyr::group_by(Vomit) %s>%
dplyr::mutate(count_Vomit = sum(count_Nausea)) %s>%
dplyr::mutate(pct = count_Nausea / count_Vomit) %s>% {
ggplot2::ggplot(., aes(x = Vomit,
y = count_Nausea,
fill = Nausea)) +
ggplot2::geom_bar(
position = "stack",
stat = "identity") +
ggplot2::geom_text(
aes(label = count_Nausea),
.,
stat = 'identity',
size = 4,
position = position_stack(vjust = 0.7, reverse = FALSE)) +
ggplot2::geom_text(
aes(label = scales::percent(pct)),
.,
stat = 'identity',
size = 4,
position = position_stack(vjust = 0.5, reverse = FALSE)) +
ggplot2::labs(.,
title = "Frequency of Nausea among Vomiting",
x = "Vomiting?",
y = "Frequency of Nausea")
}
## `summarise()` has grouped output by 'Vomit'. You can override using the `.groups` argument.
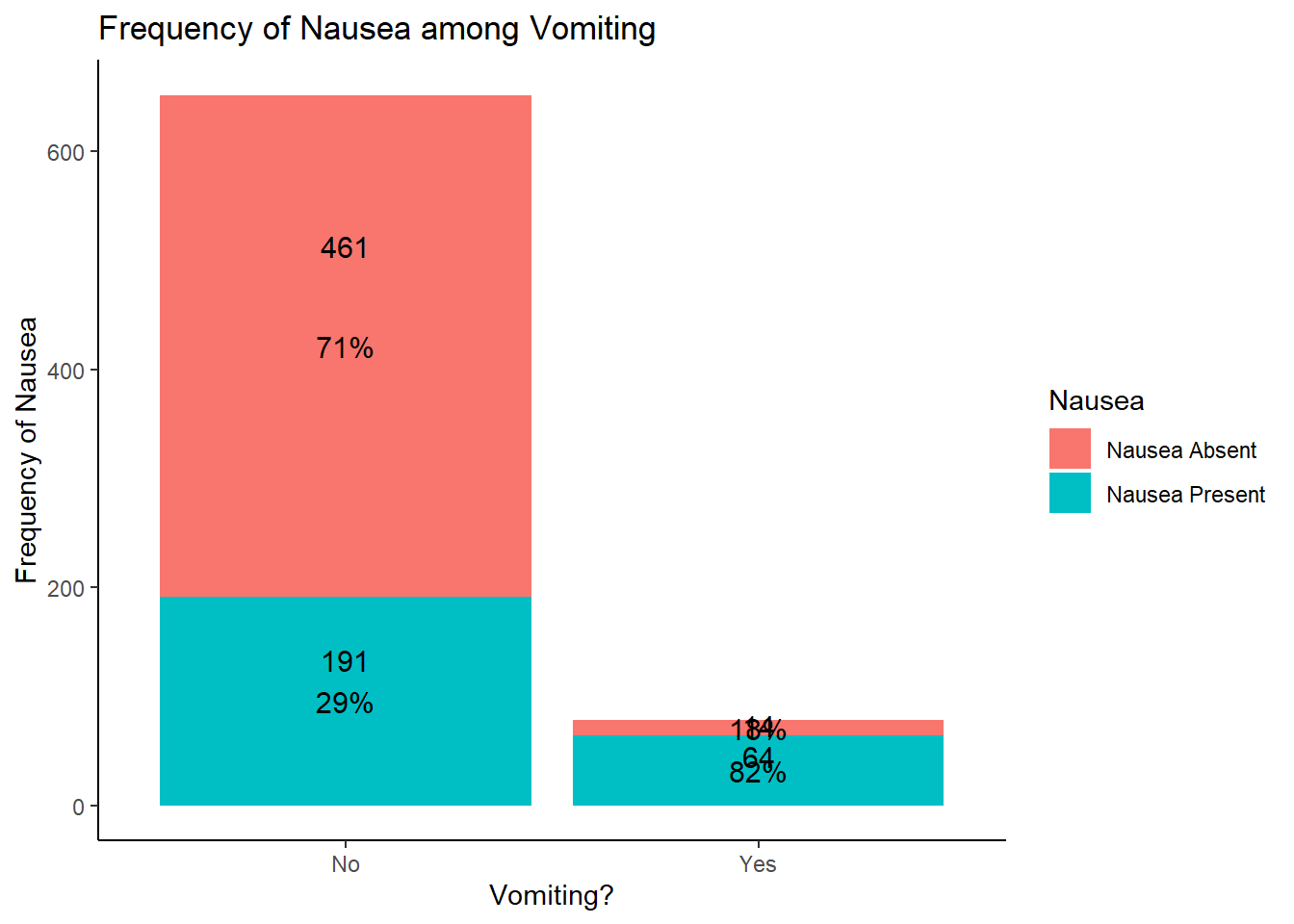
# based on the results, it appears that more patients with vomiting had nausea
# this makes sense clinically as nausea and vomiting often co-present
Creating A “Table 1” For Categorical Outcome of Interest (Nausea)
Often the first table of a manuscript lists the predictors on each row with columns representing the outcome in question. The table1 package works extremely well for categorical variables.
# first, create the summary statistics within the Table1 package for predictor variables
# already created earlier, but placed here for reference
table1::label(EDAdata$RunnyNose) <- "Runny Nose"
table1::label(EDAdata$Pharyngitis) <- "Pharyngitis"
table1::label(EDAdata$NasalCongestion) <- "Nasal Congestion"
table1::label(EDAdata$CoughIntensity) <- "Cough Intensity"
table1::label(EDAdata$ChillsSweats) <- "Chills / Sweating"
table1::label(EDAdata$Myalgia) <- "Myalgia"
table1::label(EDAdata$Vomit) <- "Vomit"
table1::label(EDAdata$Diarrhea) <- "Diarrhea"
# now, load all into a table 1 where columns represent nausea categories
table1::table1(~ RunnyNose + Pharyngitis + NasalCongestion + CoughIntensity + ChillsSweats + Myalgia + Vomit + Diarrhea
| Nausea, data = EDAdata)
|
Nausea Absent
(N=475) |
Nausea Present
(N=255) |
Overall
(N=730) |
| Runny Nose |
|
|
|
| No |
139 (29.3%) |
72 (28.2%) |
211 (28.9%) |
| Yes |
336 (70.7%) |
183 (71.8%) |
519 (71.1%) |
| Pharyngitis |
|
|
|
| No |
80 (16.8%) |
39 (15.3%) |
119 (16.3%) |
| Yes |
395 (83.2%) |
216 (84.7%) |
611 (83.7%) |
| Nasal Congestion |
|
|
|
| No |
120 (25.3%) |
47 (18.4%) |
167 (22.9%) |
| Yes |
355 (74.7%) |
208 (81.6%) |
563 (77.1%) |
| Cough Intensity |
|
|
|
| None |
30 (6.3%) |
17 (6.7%) |
47 (6.4%) |
| Mild |
99 (20.8%) |
55 (21.6%) |
154 (21.1%) |
| Moderate |
232 (48.8%) |
125 (49.0%) |
357 (48.9%) |
| Severe |
114 (24.0%) |
58 (22.7%) |
172 (23.6%) |
| Chills / Sweating |
|
|
|
| No |
103 (21.7%) |
27 (10.6%) |
130 (17.8%) |
| Yes |
372 (78.3%) |
228 (89.4%) |
600 (82.2%) |
| Myalgia |
|
|
|
| None |
63 (13.3%) |
16 (6.3%) |
79 (10.8%) |
| Mild |
159 (33.5%) |
54 (21.2%) |
213 (29.2%) |
| Moderate |
198 (41.7%) |
127 (49.8%) |
325 (44.5%) |
| Severe |
55 (11.6%) |
58 (22.7%) |
113 (15.5%) |
| Vomit |
|
|
|
| No |
461 (97.1%) |
191 (74.9%) |
652 (89.3%) |
| Yes |
14 (2.9%) |
64 (25.1%) |
78 (10.7%) |
| Diarrhea |
|
|
|
| No |
435 (91.6%) |
196 (76.9%) |
631 (86.4%) |
| Yes |
40 (8.4%) |
59 (23.1%) |
99 (13.6%) |
# in a real analysis, we could also use this table for our univariate analysis, so we could conduct a chi-square test
# to test for differences in each variable across the Nausea strata.

















